Brain Tumor Genomcis
Single-cell viral oncogenesis
Ashish H. Shah (PI)
Ashish H. Shah (PI)
A study of single-cell RNA-Seq to understand activation of antiviral immune response in glioblastoma and its role in potentiating immune checkpoint inhibition.
Single-cell transcriptiome of glioma stem cells
Luis F. Parada (PI)
Luis F. Parada (PI)
A study of single-cell RNA-Seq in order to identify gene signature of glioblastoma cancer stem cell, and identify potential new targets for therapy.
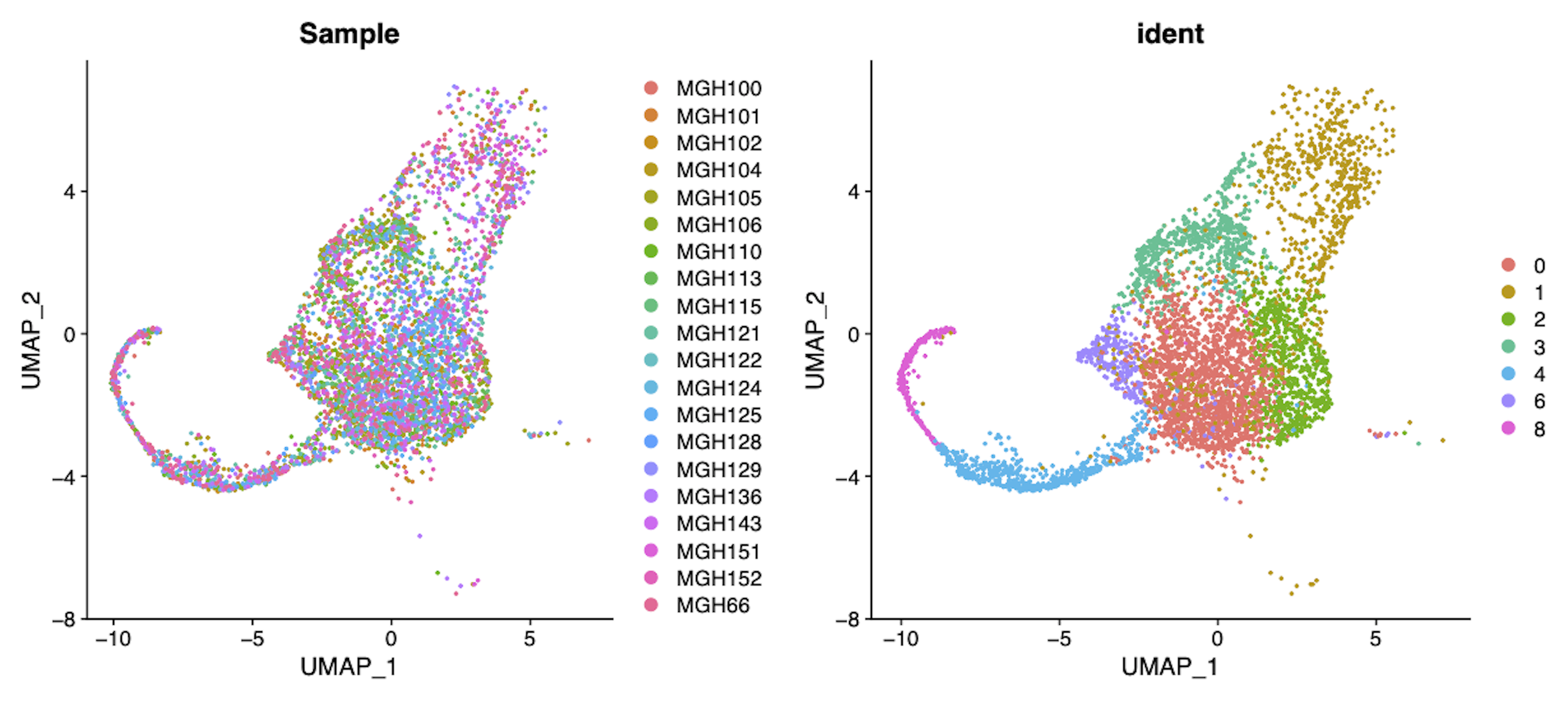
Single-cell RNA-Seq identifies 7 distinct cell types across 20 adult GBM tumors (Data from Neftel, 2019)
A study of single-cell RNA-Seq in order to identify gene signature of glioblastoma cancer stem cell, and identify potential new targets for therapy.
Single-cell multiomic profile of pituitary adenomas
Prashant Chittiboina (PI)
Prashant Chittiboina (PI)
A study of epigenomics of pituitary adenomas and normal pituitary cells to identify transcriptional regulation mechanisms responsible for pathogenesis of spontaneous pituitary adenomas.
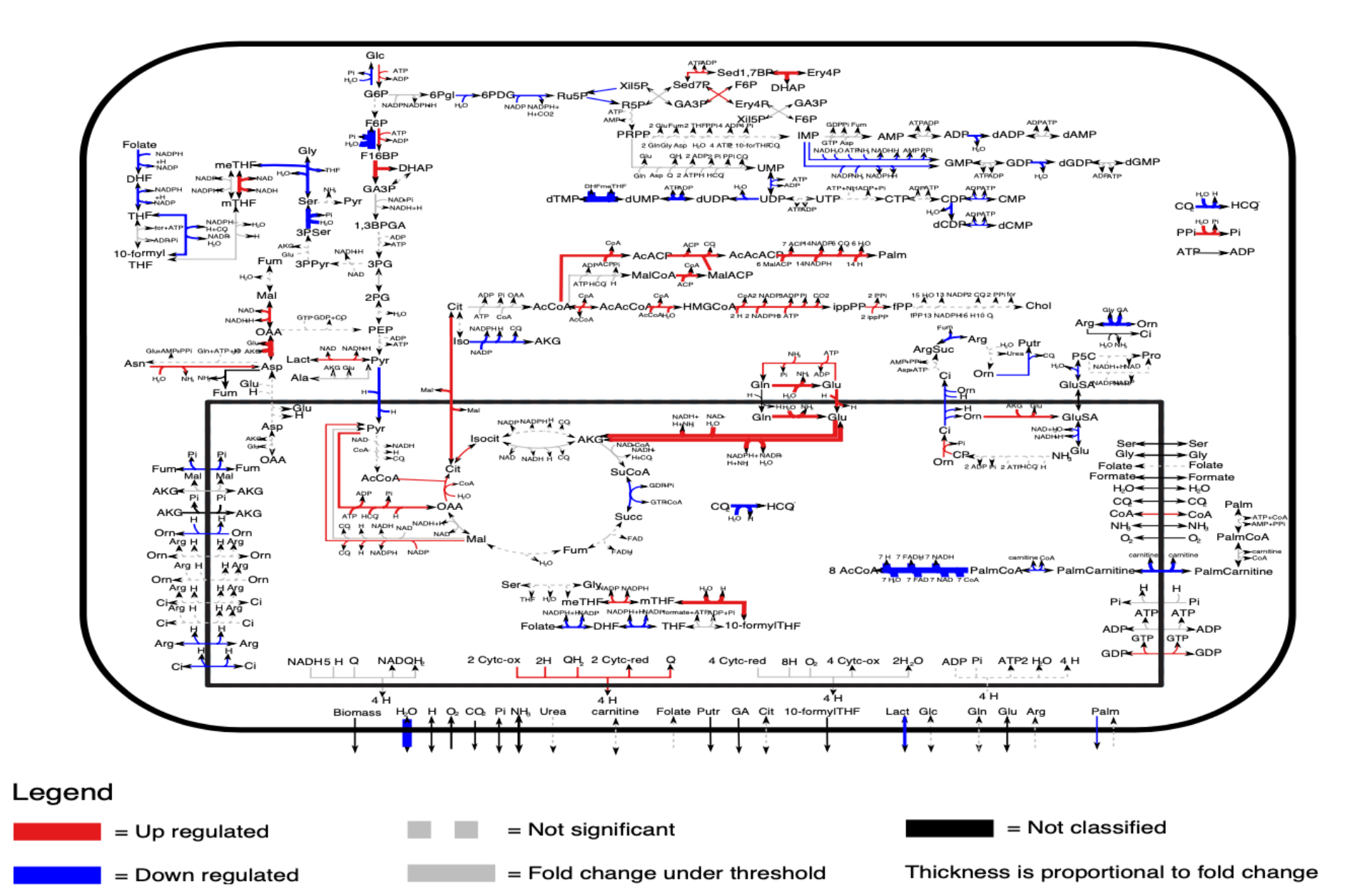
Differentially activated metabolic pathways across different tumors (Data form TCGA)
Metabolic pathways of low grade gliomas
Jack Goodman (collaborator)
Jack Goodman (collaborator)
We use metabolic pathway reactivity scores from transcriptiome sequencing data to identify low grade glioma tumors associated with prolonged patient survival.
Metablic pathway classification of tumors and prognosis of survival
Jack Goodman (collaborator)
Jack Goodman (collaborator)
We use survival modeling based on differential metabolic pathway activation to offer novel classificaiton system for tumors across all tissue types and use metabolic classification of tumors to identify patients with prolonged survival.
